The research interests of our lab encompass epigenetics, gene expression, and nuclear organization. Specifically, we focus on X chromosome inactivation and reactivation, spatial control of gene expression, and the role of long non-coding RNAs at both the single gene and genome-wide levels. Additionally, our lab is actively involved in small molecule and genetic screens, drug discovery, and the study of sex-biased diseases.
The human X-chromosome carries approximately 20-30% of genes associated with intellectual disabilities. Mutations in X-chromosome genes contribute to up to 20% of Autism Spectrum Disorders (ASDs). Epimutations on the X-chromosome have been linked to various mental health conditions, including depression, bipolar disorder, schizophrenia, and neurodegeneration.
In female mammals, one of the two X chromosomes is silenced to achieve dosage compensation with males through random X chromosome inactivation. As a result, females with an X-linked mutation typically silence both the mutated and wild-type (WT) copies of the gene, usually in a 50-50 ratio. Consequently, females possess a natural “reservoir” of non-mutated dormant genes that can potentially be reactivated.
Reactivation of these "dormant" X-linked genes is achievable through drug treatment that deactivates the master regulator of X inactivation, the long non-coding RNA (lncRNA) Xist.
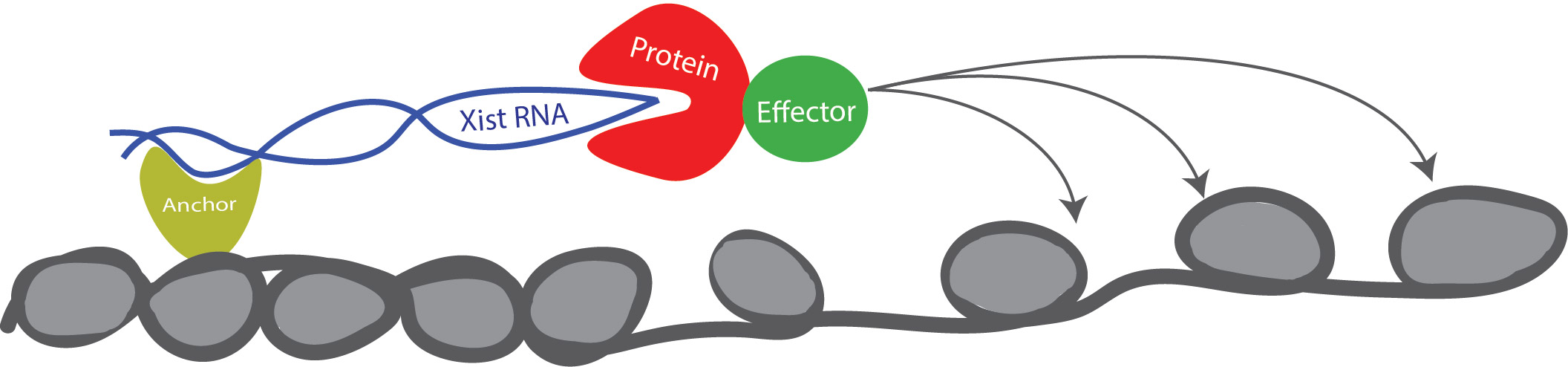
Fig.1) Xist works a molecular scaffold to recruit proteins and induce gene-silencing of an entire chromosome (one of the two X chromosomes). Blocking the interaction of Xist with silencing partners can weaken its function and reactivate genes on the inactive X-chromosome.
The reactivation of dormant genes has the potential to normalize protein levels and lead to the rescue of functional phenotypes. Our current focus involves identifying small molecules that can specifically block Xist RNA interaction with silencing partners or other Xist-specific functions.We are actively screening for compounds that can reactivate the silent wild-type copy of MeCP2 and Cdkl5. Mutations in these genes are causative of Rett and CDKL5 syndromes, respectively, both of which are debilitating ASD/intellectual-retardation diseases. Importantly, diseases associated with mutations in these genes do not lead to neurodegeneration, making them potentially curable in the post-natal/adult state, as demonstrated by previous research.
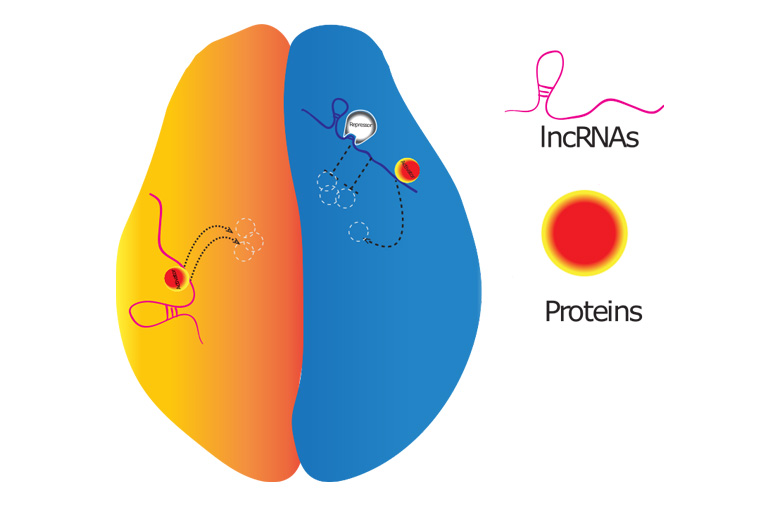
Fig. 2 LncRNAs can interact with activators (arrows) or repressors (T-lines) to activate or silence genes, respectively.
The goal of this research is to identify novel candidate long non-coding RNAs (lncRNAs) that may regulate the expression of protein-coding genes, thereby influencing brain development and function. A primary focus will be on analyzing lncRNAs associated with diseases during brain development, as well as their responsiveness to environmental factors such as stress, diet, chemicals, and aging.
Role of lncRNAs in phase separation, aggregation and in neurodegenerative disorders
Long non-coding RNAs (lncRNAs) are RNA molecules longer than 200 bases that lack coding potential. RNA contributes to the formation of membrane-less organelles, such as paraspeckles and stress granules. Xist, the master regulator of X chromosome inactivation (XCI), induces large-scale heterochromatinization of the entire X-chromosome by accumulating in large granule-like assemblies.
We recently proposed that Xist stabilizes XCI by recruiting RNA-binding proteins through a mechanism of liquid-liquid phase separation (Cerase et al., 2019/22). Xist RNA contains six conserved repetitive regions, named A to F, which are reported to be essential for its function. These regions sustain most interactions with proteins (Pintacuda et al, 2017). Specifically, these regions interact with intrinsically disordered proteins, crucial for phase separation (Cerase et al, 2019).
Our objectives include creating inducible knockdown (KD) lines of Xist-interacting proteins in mouse embryonic stem cells (mESCs) to determine the molecules required per cell to form a granule. Additionally, we plan to establish inducible Xist deletions using CRISPR/Cas9 technology to identify the regions of Xist necessary for the process. The data obtained will be used to simulate granule formation in wild-type (WT) and perturbed cells.
We aim to generate simple yet general mathematical models applicable to RNA-driven phase separation. This approach holds broad relevance, especially in the context of anti-aggregation drug development for pathologies such as neurodegeneration.
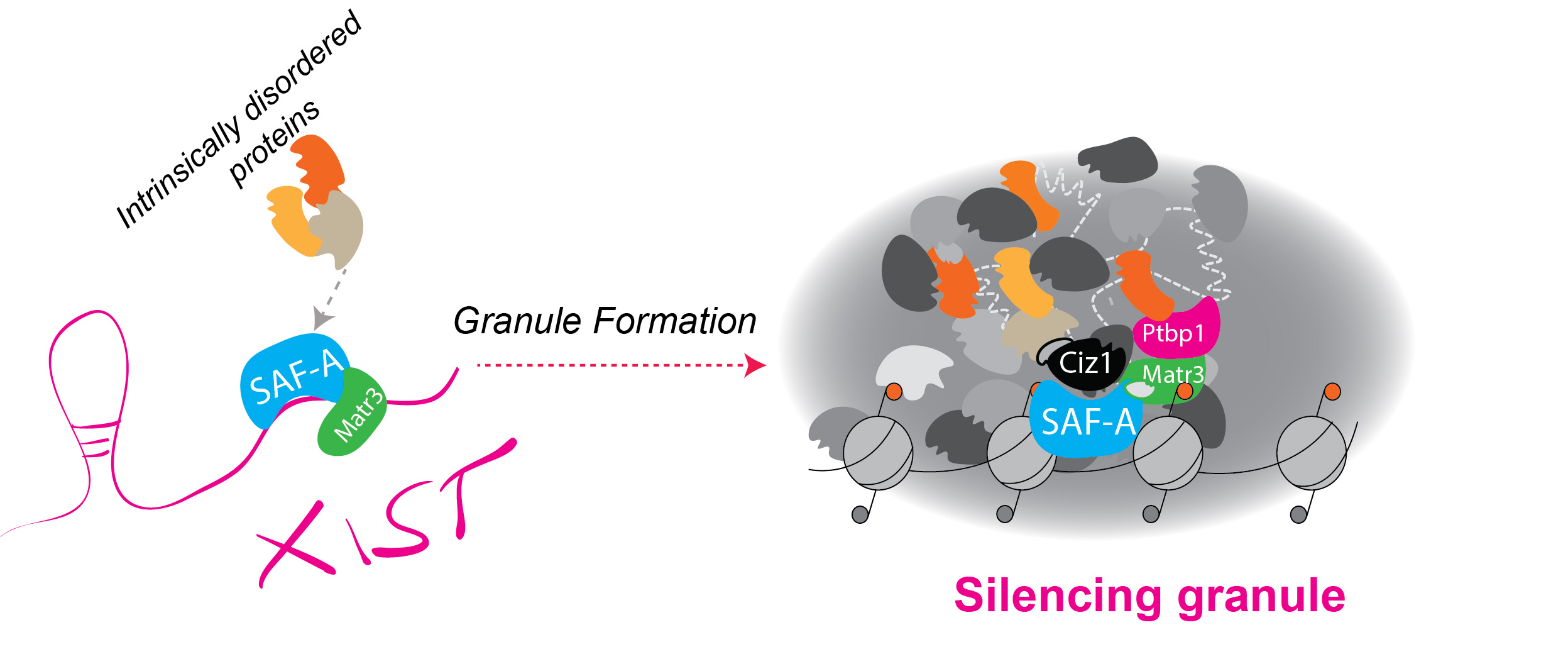
Fig. 3 Xist RNA ascts as a scaffold for protein recruitment and RNA-mediated phase separation.
With this research we aim to understand the biophysical principles regulating RNA-mediated phase separation
Functional analysis of lncRNA coding for micropeptides
While long noncoding RNAs (lncRNAs) represent the majority of RNA transcripts, their function remains elusive compared to mRNAs. Although they are known to play a crucial role in gene expression regulation and contribute to shaping the nucleus's architecture, the full spectrum of their functions is still under exploration. Complicating matters is their noncoding nature, which can be deceptive, as they sometimes code for rare, tissue-specific peptides that are challenging to detect.
Current bioinformatics tools for predicting coding potential rely on empirical user-defined parameters and exhibit limited predictive power. The objective of this project is to develop a bioinformatic prediction tool using mathematical probability theory. The tool aims to accurately identify lncRNAs that code for peptides. Subsequently, the tool's predictions will be validated by detecting these peptides through mining proteomic databases.
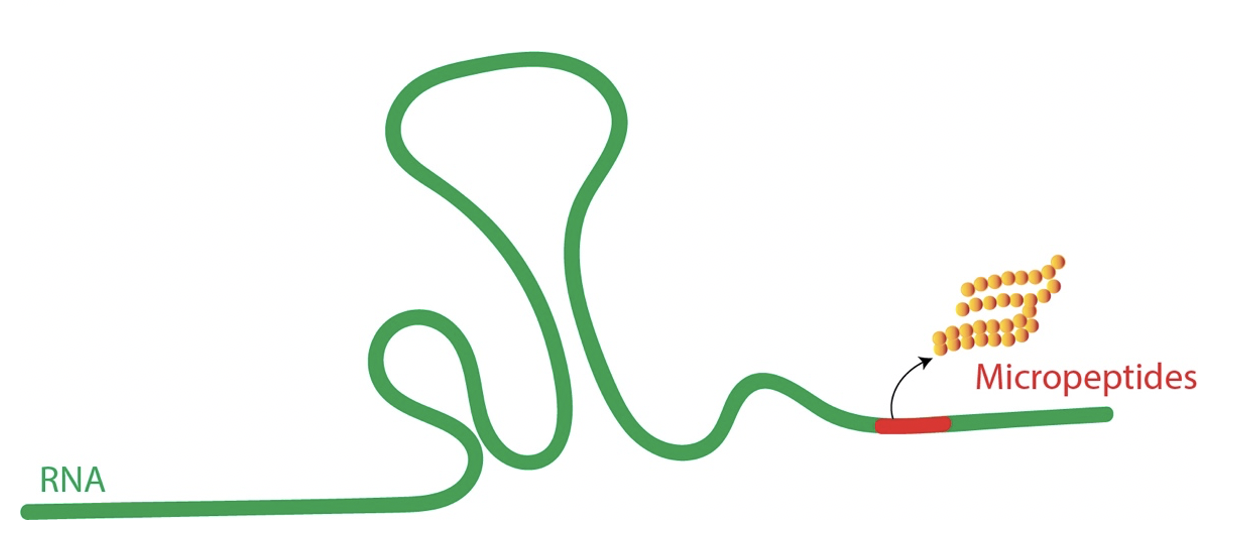
Fig.4 Bona fide lncRNAs can code for functional small peptides.
With this reserach we aim to unravel the complexity of lncRNA coding and regulatory repertoire.
Cerase lab is a part of QMUL Epigentics Hub
Teaching: Molecular Biology, Biotechnology for the Neurosciences @UnuPI; PBLs in Human Development and Neurobiology, Genomics, Epigenetics, lncRNAs, Molecular Biology @QMUL. Curriculum development: Molecular Biology, Epigenetics, X-chromosome Inactivation (in progress).
